JSOTutorials.jl
In this tutorial, you will learn how to use JSO-compliant solvers to solve a PDE-constrained optimization problem discretized with PDENLPModels.jl.
\toc
Problem Statement
In this first part, we define a distributed Poisson control problem with Dirichlet boundary conditions which is then automatically discretized. We refer to Gridap.jl for more details on modeling PDEs and PDENLPModels.jl for PDE-constrained optimization problems.
Let $\Omega = (-1,1)^2$, we solve the following problem: \(\begin{aligned} \min_{y \in H^1_0, u \in H^1} \quad & \frac{1}{2 \alpha} \int_\Omega |y(x) - y_d(x)|^2dx + \frac{\alpha}{2} \int_\Omega |u|^2dx \\ \text{s.t.} & -\Delta y = h + u, \quad x \in \Omega, \\ & y = 0, \quad x \in \partial \Omega, \end{aligned}\) where $y_d(x) = -x_1^2$ and $\alpha = 10^{-2}$. The force term is $h(x_1, x_2) = - sin(\omega x_1)sin(\omega x_2)$ with $\omega = \pi - \frac{1}{8}$.
using Gridap, PDENLPModels
# First, we define the domain and its discretization.
n = 100
domain = (-1, 1, -1, 1)
partition = (n, n)
model = CartesianDiscreteModel(domain, partition)
# Then, we introduce the definition of the finite element spaces.
reffe = ReferenceFE(lagrangian, Float64, 1)
Xpde = TestFESpace(model, reffe; conformity = :H1, dirichlet_tags = "boundary")
y0(x) = 0.0
Ypde = TrialFESpace(Xpde, y0)
reffe_con = ReferenceFE(lagrangian, Float64, 1)
Xcon = TestFESpace(model, reffe_con; conformity = :H1)
Ycon = TrialFESpace(Xcon)
Y = MultiFieldFESpace([Ypde, Ycon])
# Gridap also requires setting the integration machinery use to define next the objective function and the constraint operator.
trian = Triangulation(model)
degree = 1
dΩ = Measure(trian, degree)
yd(x) = -x[1]^2
α = 1e-2
function f(y, u)
∫(0.5 / α * (yd - y) * (yd - y) + 0.5 * α * u * u) * dΩ
end
ω = π - 1 / 8
h(x) = -sin(ω * x[1]) * sin(ω * x[2])
function res(y, u, v)
∫(∇(v) ⊙ ∇(y) - v * u - v * h) * dΩ
end
op = FEOperator(res, Y, Xpde)
npde = Gridap.FESpaces.num_free_dofs(Ypde)
ncon = Gridap.FESpaces.num_free_dofs(Ycon)
x0 = zeros(npde + ncon);
Overall, we built a GridapPDENLPModel, which implements the NLPModel API.
nlp = GridapPDENLPModel(x0, f, trian, Ypde, Ycon, Xpde, Xcon, op, name = "Control elastic membrane")
using NLPModels
(get_nvar(nlp), get_ncon(nlp))
(20002, 9801)
Find a Feasible Point
Before solving the previously defined model, we will first improve our initial guess.
The first step is to create a nonlinear least-squares whose residual is the equality-constraint of the optimization problem.
We use FeasibilityResidual from NLPModelsModifiers.jl to convert the NLPModel as an NLSModel.
Then, using trunk, a matrix-free solver for least-squares problems implemented in JSOSolvers.jl, we find an
improved guess which is close to being feasible for our large-scale problem.
By default, JSO-compliant solvers use get_x0(nlp) as an initial guess.
using JSOSolvers, NLPModelsModifiers
nls = FeasibilityResidual(nlp)
stats_trunk = trunk(nls, max_time = 300.0)
"Execution stats: first-order stationary"
We check the solution from the stats returned by trunk:
norm(cons(nlp, stats_trunk.solution))
4.465311648679671e-7
We will use the solution found to initialize our solvers.
Solve the Problem
Finally, we are ready to solve the PDE-constrained optimization problem with a targeted tolerance of 10⁻⁵.
In the following, we will use both Ipopt and DCI on our problem.
We refer to the tutorial How to solve a small optimization problem with Ipopt + NLPModels
for more information on NLPModelsIpopt.
using NLPModelsIpopt
# Set `print_level = 0` to avoid printing detailed iteration information.
stats_ipopt = ipopt(nlp, x0 = stats_trunk.solution, tol = 1e-5, print_level = 0)
******************************************************************************
This program contains Ipopt, a library for large-scale nonlinear optimization.
Ipopt is released as open source code under the Eclipse Public License (EPL).
For more information visit https://github.com/coin-or/Ipopt
******************************************************************************
"Execution stats: first-order stationary"
The problem was successfully solved, and we can extract the function evaluations from the stats.
nlp.counters
Counters:
obj: █⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅ 6 grad: █⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅ 7 cons: █⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅ 13
cons_lin: ⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅ 0 cons_nln: █⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅ 13 jcon: ⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅ 0
jgrad: ⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅ 0 jac: █⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅ 7 jac_lin: ⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅ 0
jac_nln: █⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅ 7 jprod: ████████████████████ 1127 jprod_lin: ⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅ 0
jprod_nln: ████████████████████ 1127 jtprod: ████████████████████ 1133 jtprod_lin: ⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅ 0
jtprod_nln: ████████████████████ 1133 hess: █⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅ 5 hprod: ⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅ 0
jhess: ⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅ 0 jhprod: ⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅ 0
Reinitialize the counters before re-solving.
reset!(nlp);
Most JSO-compliant solvers are using logger for printing iteration information.
NullLogger avoids printing iteration information.
using DCISolver, Logging
stats_dci = with_logger(NullLogger()) do
dci(nlp, stats_trunk.solution, atol = 1e-5, rtol = 0.0)
end
"Execution stats: first-order stationary"
The problem was successfully solved, and we can extract the function evaluations from the stats.
nlp.counters
Counters:
obj: █⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅ 7 grad: █⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅ 7 cons: █⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅ 7
cons_lin: ⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅ 0 cons_nln: █⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅ 7 jcon: ⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅ 0
jgrad: ⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅ 0 jac: █⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅ 13 jac_lin: ⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅ 0
jac_nln: █⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅ 13 jprod: ████████████████████ 10895 jprod_lin: ⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅ 0
jprod_nln: ⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅ 0 jtprod: ████████████████████ 10895 jtprod_lin: ⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅ 0
jtprod_nln: ⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅ 0 hess: █⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅ 5 hprod: ⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅ 0
jhess: ⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅ 0 jhprod: ⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅⋅ 0
We now compare the two solvers with respect to the time spent,
stats_ipopt.elapsed_time, stats_dci.elapsed_time
(74.27, 20.94061589241028)
and also check objective value, feasibility, and dual feasibility of ipopt and dci.
(stats_ipopt.objective, stats_ipopt.primal_feas, stats_ipopt.dual_feas),
(stats_dci.objective, stats_dci.primal_feas, stats_dci.dual_feas)
((16.044043741825803, 2.740863092043355e-16, 6.754773183374242e-6), (16.040271504407357, 5.628478048695899e-10, 7.394013205487982e-8))
Overall DCISolver is doing great for solving large-scale optimization problems!
You can try increase the problem size by changing the discretization parameter n.
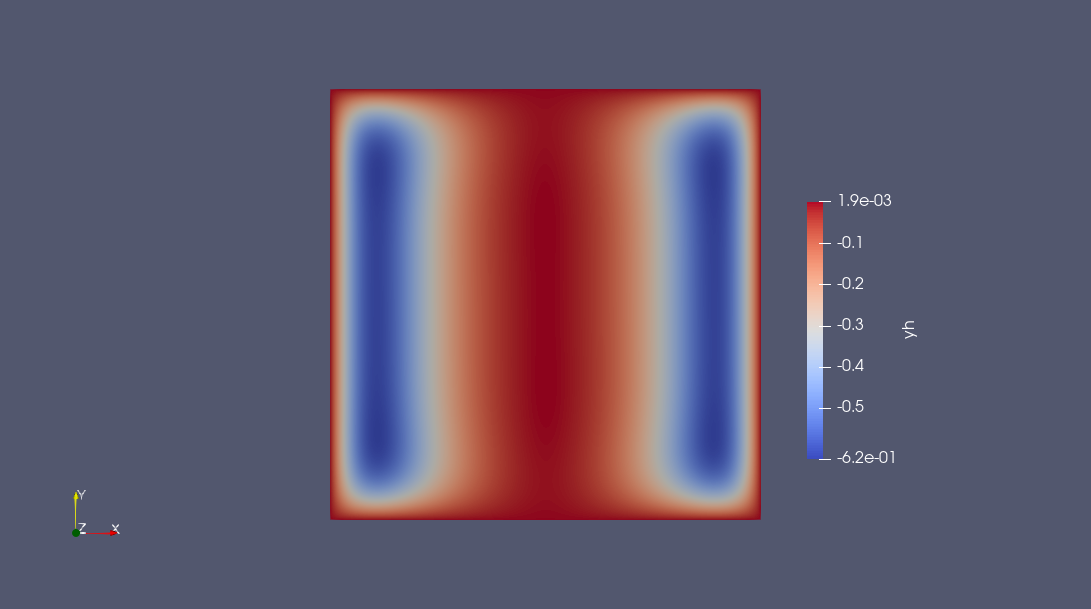
Finally, switching the discrete solution as a FEFunction the result can written as a VTK-file using Gridap’s facilities.
yfv = stats_dci.solution[1:Gridap.FESpaces.num_free_dofs(nlp.pdemeta.Ypde)]
yh = FEFunction(nlp.pdemeta.Ypde, yfv)
ufv = stats_dci.solution[1+Gridap.FESpaces.num_free_dofs(nlp.pdemeta.Ypde):end]
uh = FEFunction(nlp.pdemeta.Ycon, ufv)
writevtk(nlp.pdemeta.tnrj.trian, "results", cellfields = ["uh" => uh, "yh" => yh])
(["results.vtu"],)
The following plots can obtained using any software reading VTK, e.g. Paraview.